Faster R package installation
Faster package installation
Every few weeks or so, a tweet pops up asking about how to speed up package installation in R

Depending on the luck of twitter, the author may get a few suggestions.
The bigger picture is that package installation time is starting to become more of an issue for a number of reasons. For example, packages are getting larger and more complex (tidyverse and friends), so installation just takes longer. Or we are using more continuous integration strategies such as Travis or GitLab-CI, and want quick feedback. Or we are simply updating a large number of packages via update.packages(). This is a problem we often solve for our clients - optimising their CI/CD pipelines.
The purpose of this blog post is to pull together a few different methods for tackling this problem. If I’ve missed any, let me know (https://twitter.com/csgillespie)!
Faster installation with Ncpus
The first tactic you should use is the Ncpus argument in install.packages() and update.packages(). This installs packages in parallel. It doesn’t speed up an individual package installs, but it does allow dependencies to install in parallel, e.g. tidyverse. Using it is easy; it’s just an additional argument in install.packages(). So to use six cores, we would simply use
install.packages("tidyverse", Ncpus = 6)
When installing a fresh version of the tidyverse and all dependencies, this can give a two-fold speed-up.
| Ncpus | Elapsed (Secs) | Ratio |
|---|---|---|
| 1 | 409 | 2.26 |
| 2 | 224 | 1.24 |
| 4 | 196 | 1.08 |
| 6 | 181 | 1.00 |
Not bad for a simple tweak with no downsides. For further information, see our blog post from a few years ago.
In short, this is something you should definitely use and add to your .Rprofile. It would in theory speed-up continuous integration pipelines, but only if you have multiple cores available. The free version of travis only comes with a single core, but if you hook up a multi-core Kubernettes cluster to your CI (we sometimes do this at Jumping Rivers), then you can achieve a large speed-up.
Faster installation with ccache
If you are installing packages from source, i.e. tar.gz files, then most of the installation time is spent on compiling source code, such as C, C++ & Fortran. A few years ago, Dirk Eddelbuettel wrote a great blog post on leveraging the ccache utility for reducing the compile time step. Essentially, ccache stores the resulting object file created when compiling. If that file is ever compiled again, instead of rebuilding, ccache returns the object code, resulting in a significant speed up. It’s the classic trade-off between memory (caching) and CPU.
Dirk’s post gives clear details on how to implement ccache (so I won’t repeat). He also compares re-installation times of packages, with RQuantlib going from 500 seconds to a few seconds. However, for ccache to be effective, the source files have to be static. Obviously, when you update an R package things change!
As an experiment, I download the last seventeen versions of {dplyr} from CRAN. This takes us back to version 0.5.0 from 2016. Next I installed each version in turn, via
# Avoid tidyverse packages, as we are messing about with dplyr
f = list.files("data", full.names = TRUE)
elapsed = numeric(length(f))
for (i in seq_along(f)) {
elapsed[i] = system.time(install.packages(f[i], repos = NULL))["elapsed"]
}
As all packages dependencies have been installed and the source code has already been downloaded, the above code times the installing of just {dplyr}. If we then implement ccache, we can easily rerun the above code. After a little manipulation we can plot the absolute installation times

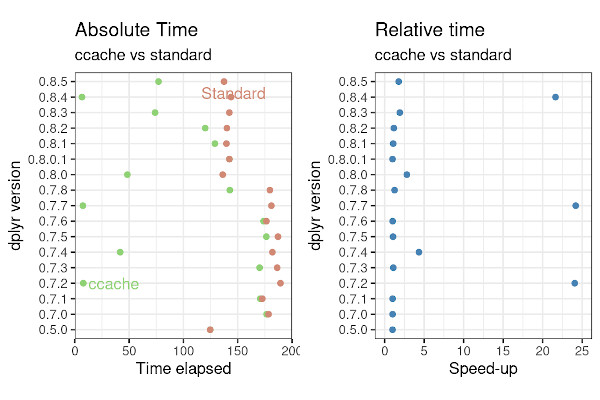
The first (slightly obvious) takeaway is that there is no speed-up with {dplyr} v0.5.0. This is simply because ccache relies on previous installations. As v0.5.0 is the first version in our study, there is no difference between standard and ccache installations.
Over the seventeen versions of dplyr, we achieved a 24 fold speed-up for three versions, and more modest two to four fold speed-up for a further three versions. Averaged over all seventeen version, a typical speed-up is around 50%.
Overall, using ccache is a very effective and easy strategy. It requires a single, simple set-up, and doesn’t require root access. Of course it doesn’t always work, but it never really slows anything down.
At the start of this section, I mentioned the trade off between memory and CPU. I’ve been using ccache since 2017, and the current cache size is around 6GB. Which on a modern hard drive isn’t much (and I install a lot of packages)!
Using Ubuntu Binaries
On Linux, the standard way of installing packages is via source and install.packages(). However, it is also possible to install packages using binary packages. This has two main benefits
- It’s faster - typically a few seconds
- It (usually) solves any horrible dependency problems by installing the necessary dev-libraries.
If you are using continuous integration, such as GitLab runners, then this is a straightforward step to reduce the package installation time. The key idea is to add an additional binary source to your source.lists file, see for example, the line in rocker. After that, you can install most CRAN packages via
sudo apt install r-cran-dplyr
The one big downside here is that the user requires root access to install an R package, so this solution isn’t suitable in all situations.
There’s lots of documentation available, CRAN and blog posts, so I won’t bother repeating by adding more.
Using RStudio Package Manager
The RStudio Package Manager is one of RStudio’s Pro products that is used to ultimately pay for their open source work, e.g. the RStudio desktop IDE and all of their tidyverse R packages.
CRAN mirrors have for a long time distributed binary packages for Windows and Mac. The RSPM provides precompiled binaries for CRAN packages for
- Ubuntu 16.04 (Xenial), Ubuntu 18.04 (Bionic)
- CentOS/RHEL 7, CentOS/RHEL 8
- openSUSE 42/SLES 12, openSUSE 15/SLES 15
- Windows (soon, currently in beta)
The big advantage of RSPM over the Ubuntu binaries solution above, is that root access is no longer necessary. Users can just install via the usual install.packages().

